Learn Genome Assembly & Annotation In Prokaryote & Eukaryote
Published 8/2025
MP4 | Video: h264, 1920x1080 | Audio: AAC, 44.1 KHz
Language: English | Size: 1.64 GB | Duration: 3h 23m
Published 8/2025
MP4 | Video: h264, 1920x1080 | Audio: AAC, 44.1 KHz
Language: English | Size: 1.64 GB | Duration: 3h 23m
Step-by-Step Guide to Assembling and Annotating Eukaryotic and Prokaryotic Genomes
What you'll learn
Understand the biological principles behind genome assembly and annotation
Set up a Linux-based bioinformatics environment, including WSL on Windows
Download and manage raw sequencing data from public repositories like NCBI and ENA
Perform quality control and trimming of sequencing reads using FastQC and porechop
Assemble genomes using SPAdes and Flye
Annotate prokaryotic genomes using Prokka
Annotate Eukaryotic Genomes using AUGUSTUS
Run BLAST for functional annotation and pathway mapping
Predict protein domains and GO terms using HMMER and Pfam databases
Visualize and validate annotation results using Quast
Build reproducible genome assembly and annotation pipelines using Bash scripting
Requirements
Basic understanding of molecular biology and genomics (e.g., DNA structure, gene expression)
Familiarity with command-line tools (Linux/Unix) is helpful but not mandatory — we’ll cover the essentials
A computer with internet access and at least 8 GB RAM (recommended for genome assembly tasks)
Ability to install and run bioinformatics tools (we’ll guide you through setting up WSL or a Linux VM)
Curiosity and willingness to learn hands-on through real datasets and practical exercises
Description
Learn Step by Step Genome Assembly & Annotation in Prokaryotes and EukaryotesTurn raw DNA sequencing data into meaningful biological insights. In this practical, hands-on bioinformatics course, you’ll learn how to perform genome assembly and annotation for both prokaryotic and eukaryotic organisms using real datasets and powerful open-source tools.Whether you’re a student, researcher, or bioinformatics enthusiast, this course will help you master skills used in genomics, biotechnology, medicine, and evolutionary biology. No prior programming experience is required. We start from the basics and guide you through each step.What You’ll LearnPerform quality control and preprocessing of sequencing readsAssemble genomes with tools like SPAdes and FlyeAnnotate genes using Prokka and AUGUSTUSAdd functional data with BLAST and PfamVisualize genomes using QuastBuild reproducible pipelines with Bash scriptingWork confidently with both prokaryotic and eukaryotic datasetsWho This Course Is ForBiology and bioinformatics studentsResearchers and lab techniciansEducators teaching genomics hands-onSelf-learners curious about genome analysisWhat You NeedJust a computer, internet connection, and basic biology knowledge. We’ll guide you through setting up your bioinformatics environment and provide downloadable scripts for practice.By the end of this course, you’ll be able to take raw sequencing reads, assemble a genome, annotate it, visualize results, and prepare them for publication or database submission.Enroll now and start your journey into genome assembly and annotation turning data into discovery.
Overview
Section 1: Course Introduction
Lecture 1 Introduction
Section 2: Linux for Bioinformatics
Lecture 2 Introduction to Linux for Bioinformatics
Lecture 3 Installing Linux or WSL on Windows
Lecture 4 Navigating the Linux File System
Lecture 5 Basic Linux Commands Every Bioinformatician Must Know
Lecture 6 Working with Files: Viewing, Editing, and Processing Genomic Data
Lecture 7 Setting Up Conda and Environment
Section 3: Short Read Genome Assembly and Analysis Using Linux
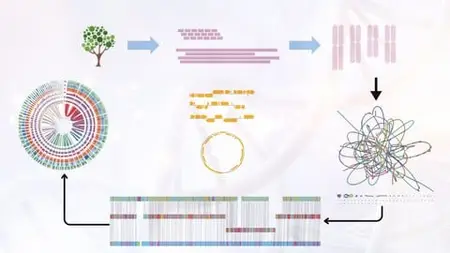
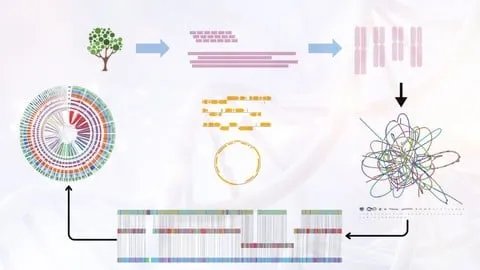
Lecture 8 Introduction to Genome Assembly Methods & Algorithms
Lecture 9 Overview of Short Read Sequencing Technologies
Lecture 10 Retrieving Short Read Data from Public Repositories
Lecture 11 Installing Tools & Performing Quality Control of Short Reads
Lecture 12 Assembling Short Reads And Visualizing Contigs And Scaffolds
Section 4: Long Read Genome Assembly and Analysis Using Linux
Lecture 13 Introduction to Long Read Sequencing Technologies
Lecture 14 Retrieving Long Read Data & Setting Up the Environment
Lecture 15 Quality Assessment of Long Reads
Lecture 16 Assembling Long Reads into Contigs and Visualizing
Section 5: Prokaryotic Genome Annotation
Lecture 17 Introduction to Genome Annotation
Lecture 18 Prokaryotic Genome Structural Annotation Using Prokka
Lecture 19 Prokaryotic Genome Functional Annotation Using BLAST
Lecture 20 Prokaryotic Genome Domain Prediction Using HMMER
Section 6: Eukaryotic Genome Annotation
Lecture 21 Eukaryotic Genome Structural Annotation
Lecture 22 Eukaryotic Genome Functional Annotation
Biology and bioinformatics students eager to gain hands-on experience in genome assembly and annotation,Early-career researchers looking to analyze sequencing data for their thesis or publications,Educators and mentors who want to integrate practical genomics workflows into their teaching,Lab technicians and data analysts working with NGS data who need reproducible pipelines,Self-learners and enthusiasts curious about how genomes are decoded, assembled, and annotated,Anyone preparing for graduate studies or research roles in genomics, computational biology, or molecular diagnostics